What is gene mapping?
Gene mapping is when we identify the locus of a gene and the distances between genes, and the map can also show the distances between different sites within a gene.
During genome mapping, a collection of molecular markers are placed onto their respective positions on the genome.
Gene mapping is often necessary before researchers can move on to more detailed identification of a gene.
The process of identifying a genetic element that is responsible for a health issue is one of the most well-known aspect of gene mapping.
Is genome mapping the same as genome sequencing?
No, it is not.
Genome mapping and genome sequencing both create a description of a genome, but genome sequencing provides a much more detailed and full description.
When a genome has been sequenced, we know the order of every DNA base in the genome. Genome mapping, on the other hand, only identifies a series of landmarks in the genome.
Example:
- Genome sequence: GCCATTGACGTCCCCTTGAAAGAGAGAGAGACTCCGATGCG
- Genome map: GCC———————CCCC———————————–CTCCG———
Even though the genome sequence contains more information than the genome map, you might still need the map to make sense of the sequence. Simply looking at the sequence is usually not enough, as it is difficult to quickly know which parts are important for your purposes. A map with landmarks can make it easier to understand where the relevant parts of the genome are.
Genome mapping is useful to researchers as it identfies and records the location of genes and the distance between certain genes within the chromosome. As mentioned above, it highlights important “landmarks” within the genome. These landmarks can be short DNA sequences, individual genes, or regulatory sites that turn genes on and off.
As research progresses, the map of a particular genome gets more and more filled in.
Mapping a genome by genetic mapping or physical mapping
The two main types of genome mapping are genetic mapping and physical mapping, and each comes with it own advantages and usefulness depending on the contex.
Both genetic mapping and physical mapping will help you find the location of a gene or a particular section of DNA in a chromosome, but the maps rely on different information.
Genetic mapping focuses on how genetic information is shuffled (either between different chromosomes or between different locations within a single chromosome) during meiosis. Meiosis is a special type of cell division.
Physical mapping focuses on the physical distance between known genes and DNA sequences, and involves knowing the number of base pairs between them. Examples of techniques used for physical mapping are fluorescent in situ hybridisation (FISH) mapping, restriction mapping (fingerprint mapping and optical mapping), and sequenced tagged site (STS) mapping.
Early genetic mapping
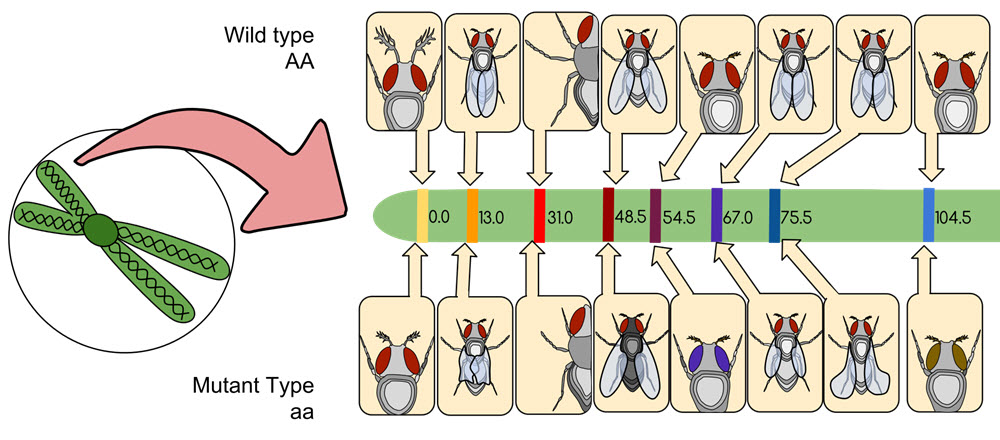
The first genetic map of a chromosome was created by Alfred Sturtevant in 1913. The chromosome was that of a Drosophila melanogaster fruit fly. Sturtevant determined that the genes in the chromosome were arranged in a linear way, and that genes for specific traits could be found in specific locations along the line.
Sturtevant was also the researcher who proposed that genes that are located close to each other on the line are more likely to be inherited together compared to genes located at a greater distance from each other. The larger the distance between two genes, the larger is the space over which recombination can happen.
What is a linkage map?
A linkage map show where the genes are in relation to each other on the chromosome. It is created by finding out how often certain characteristic are inherited together, as this information can be used to estimate the distance.
The distance between genes on the same chromosome is called the linkage distance. The shorter the linkage distance, the larger the chance of them being inherited together.
Example: Blue eyes and blond hair are inherited traits that frequently are inherited together. Therefore, we can assume that the linkage distance between the involved genes is short.
How are modern genetic maps created?
The most modern genetic maps we have were created using isolated DNA taken from blood or tissue samples of closely related individuals from a family where some exhibit a certain trait (e.g. a certain genetic health issue). Using this technique, researchers have been able to work out the position of genes by first finding out the exact frequency of genetic recombination.
The method relies on researchers carefully examining the DNA in search of unique patterns visible only in samples taken from individuals that exhibit the specific trait. These patterns are known as markers, and they can help us track how traits are inherited over many generations.
If a particular gene is close to a marker, it is more likely to be inherited together with the marker, as they are more likely to stay together during the recombination process.
